An annotated SAM format example
SAM (Sequence Alignment/Map) format is one of the most popular formats used for storing aligment information of high-throughput sequencing data. I’ve used it extensively, but still sometimes I find it hard to recognize the columns at first glance. So I annotated an example from the SAM format specification (Version: 1 Jun 2017) for quick reference.
Other related specification can be found at http://samtools.github.io/hts-specs/.
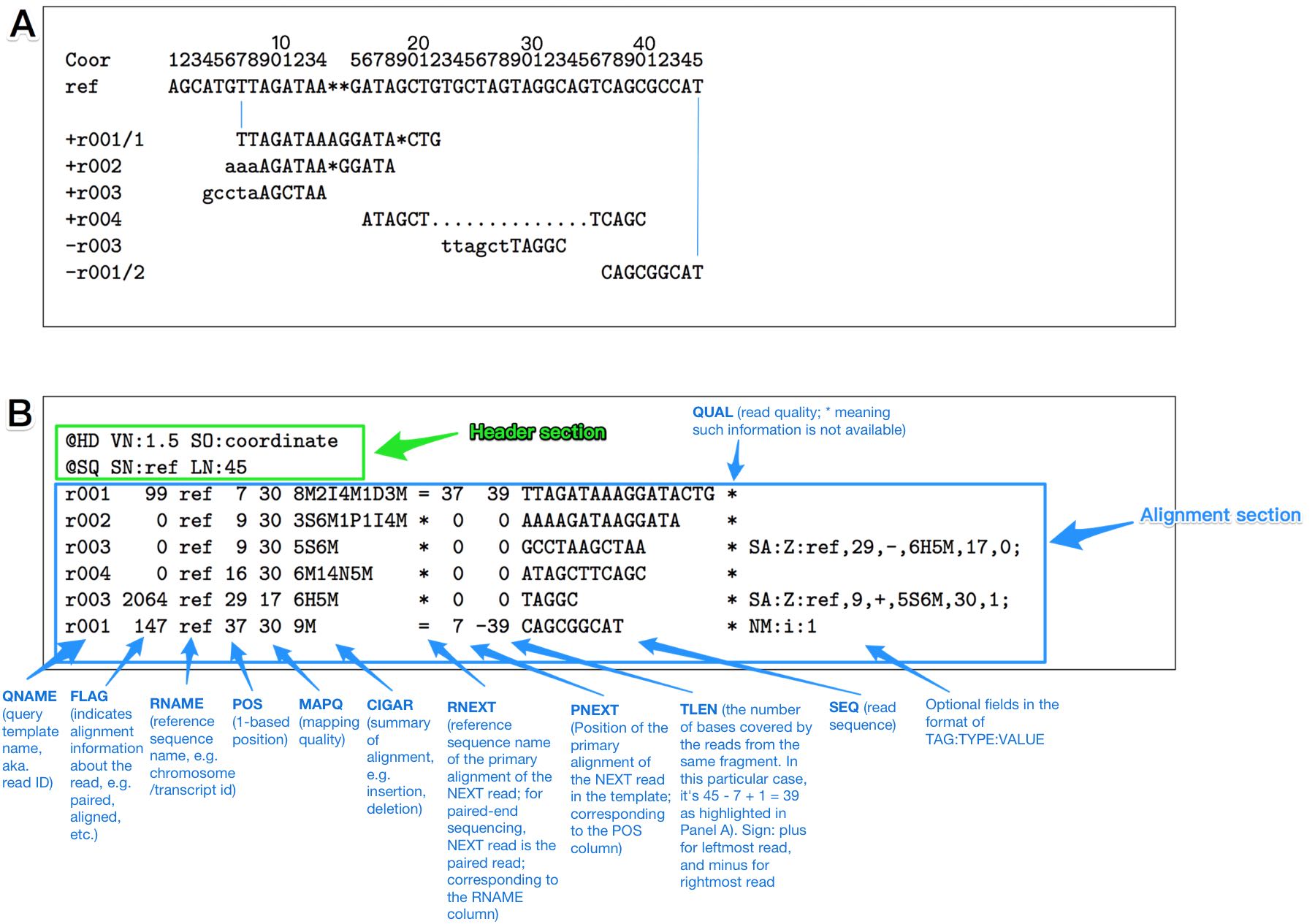
An annotated example of SAM format. (A) An example alignment result. (B) The alignment result represented in SAM format. Black color represents the SAM information, and colorful text is the annotation.